已解决
PSP - 蛋白质复合物结构预测 Template Pair 特征 Mask 可视化
来自网友在路上 182882提问 提问时间:2023-11-11 14:45:18阅读次数: 82
最佳答案 问答题库828位专家为你答疑解惑
欢迎关注我的CSDN:https://spike.blog.csdn.net/
本文地址:https://spike.blog.csdn.net/article/details/134333419
在蛋白质复合物结构预测中,在 TemplatePairEmbedderMultimer 层中 ,构建 Template Pair 特征的源码,即:
- 将特征
template_dgram、pseudo_beta_mask_2d、aatype_one_hot、backbone_mask_2d、unit_vector(x/y/z)特征,通过 linear 层累加到一起。 - 其中,都需要使用
multichain_mask_2d进行固定掩码,选择单链区域。 - 输出维度:
([1, 1102, 1102, 64]),linear层的输出c_out维度是 64。
源码如下:
def forward(self,template_dgram: torch.Tensor,aatype_one_hot: torch.Tensor,query_embedding: torch.Tensor,pseudo_beta_mask: torch.Tensor,backbone_mask: torch.Tensor,multichain_mask_2d: torch.Tensor,unit_vector: geometry.Vec3Array,
) -> torch.Tensor:act = 0.0pseudo_beta_mask_2d = (pseudo_beta_mask[..., None] * pseudo_beta_mask[..., None, :])pseudo_beta_mask_2d = pseudo_beta_mask_2d * multichain_mask_2dtemplate_dgram = template_dgram * pseudo_beta_mask_2d[..., None]act += self.dgram_linear(template_dgram)act += self.pseudo_beta_mask_linear(pseudo_beta_mask_2d[..., None])aatype_one_hot = aatype_one_hot.to(template_dgram.dtype)act += self.aatype_linear_1(aatype_one_hot[..., None, :, :])act += self.aatype_linear_2(aatype_one_hot[..., None, :])backbone_mask_2d = backbone_mask[..., None] * backbone_mask[..., None, :]backbone_mask_2d = backbone_mask_2d * multichain_mask_2dx, y, z = [coord * backbone_mask_2d for coord in unit_vector]act += self.x_linear(x[..., None])act += self.y_linear(y[..., None])act += self.z_linear(z[..., None])act += self.backbone_mask_linear(backbone_mask_2d[..., None])query_embedding = self.query_embedding_layer_norm(query_embedding)act += self.query_embedding_linear(query_embedding)return act
template_dgram 特征:
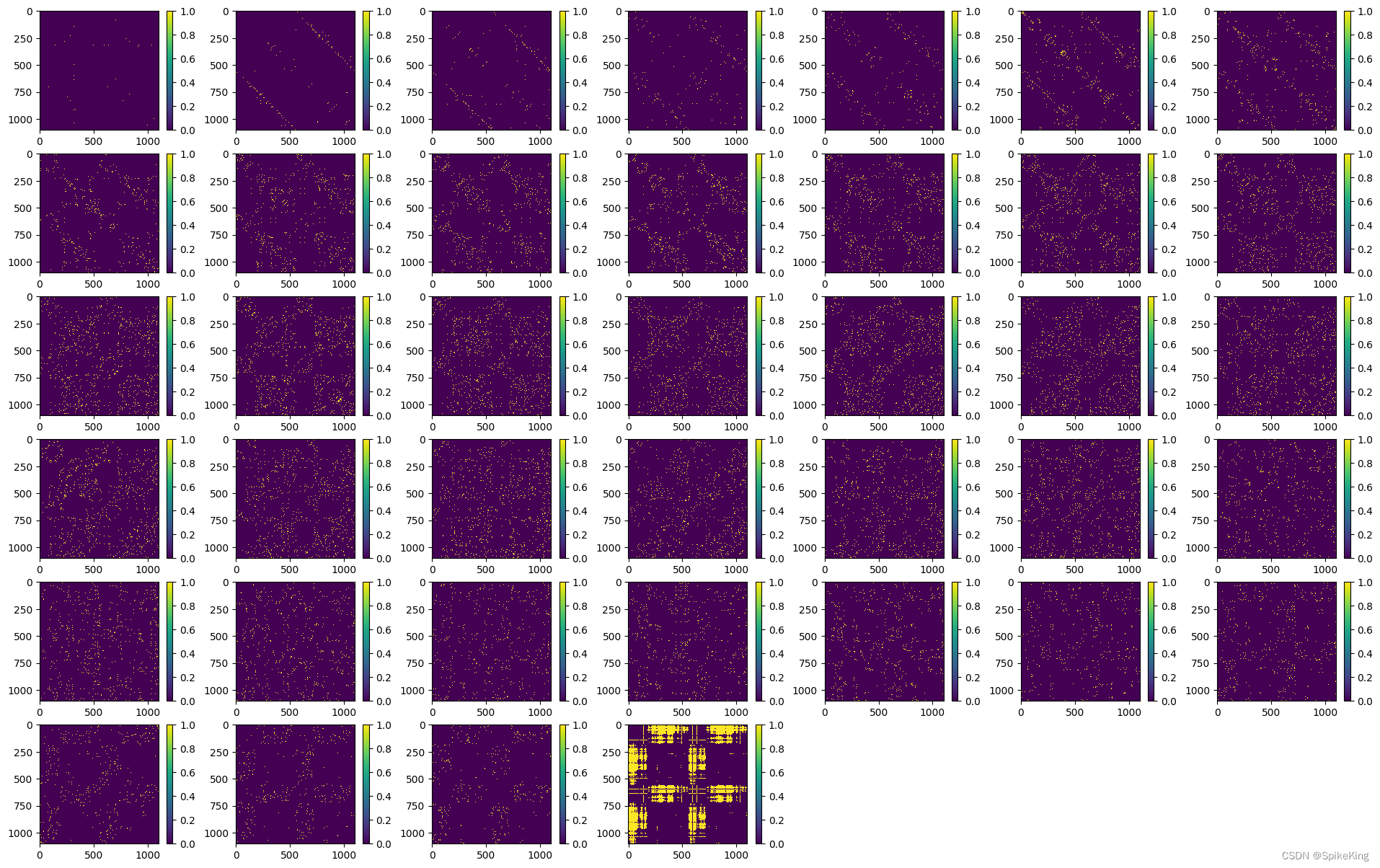
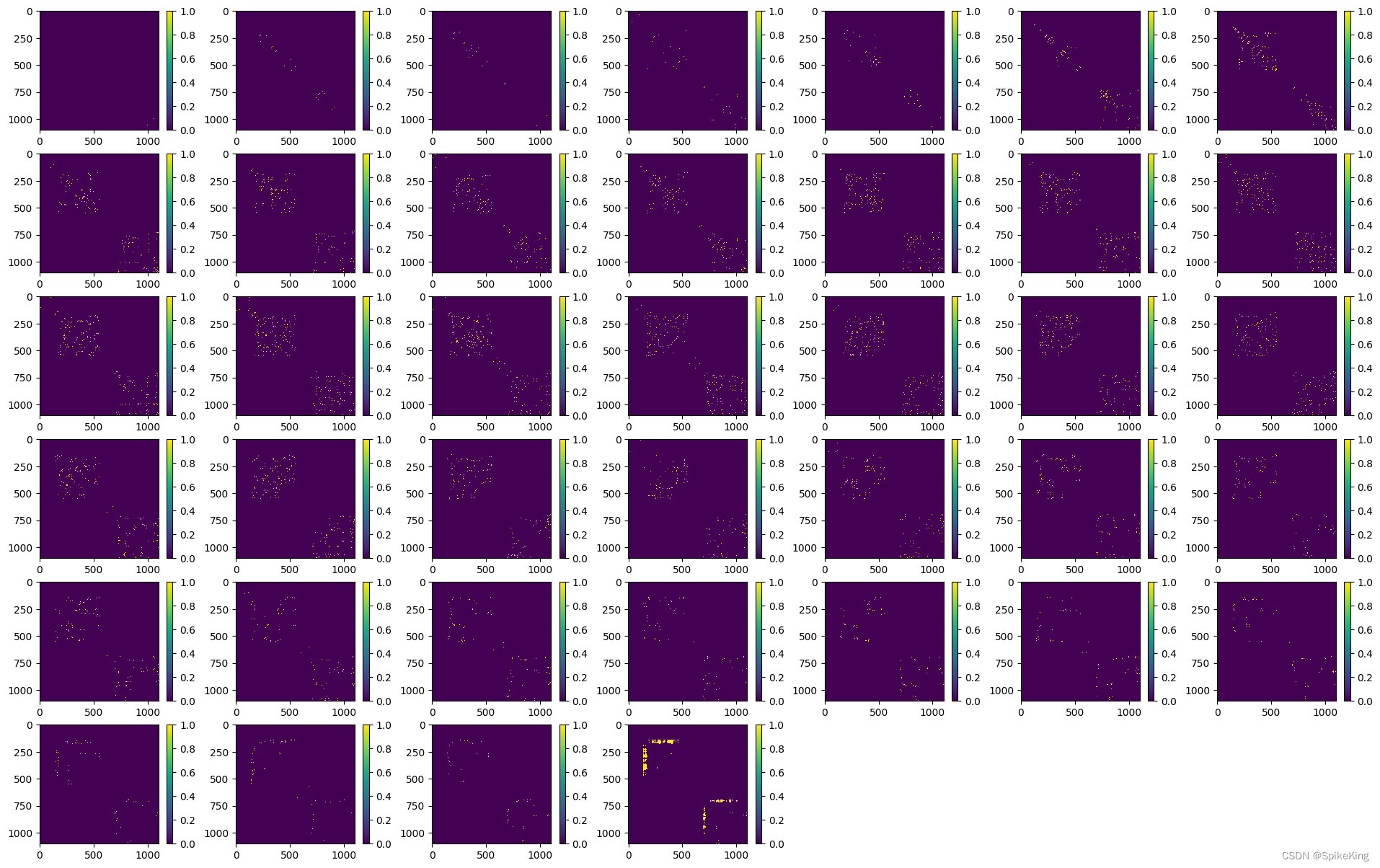
template_dgram 特征与 multichain_mask_2d:
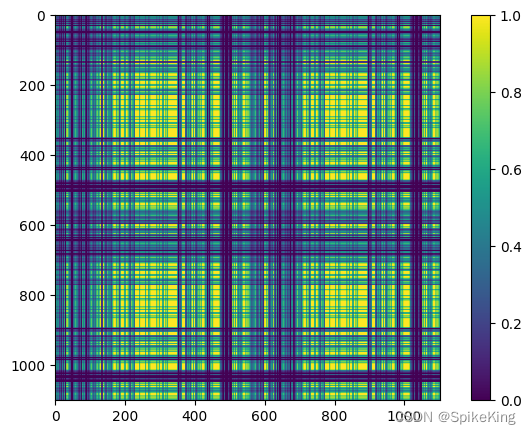
backbone_mask_2d 特征:
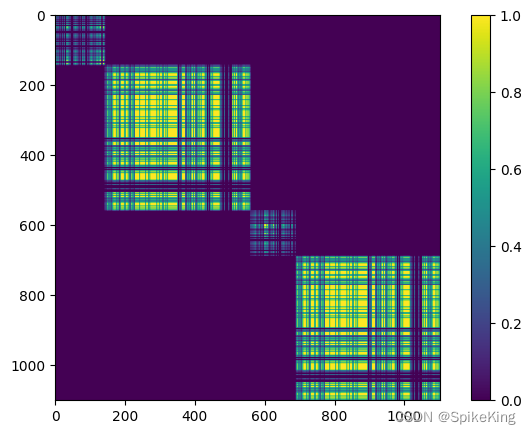
backbone_mask_2d 特征与 multichain_mask_2d:
写入特征,即:
tmp_dict = dict()
tmp_dict["pseudo_beta_mask_2d_prev"] = pseudo_beta_mask_2d.cpu().numpy()
tmp_dict["pseudo_beta_mask_2d_post"] = pseudo_beta_mask_2d.cpu().numpy()
tmp_dict["template_dgram_post"] = template_dgram.cpu().numpy()
tmp_dict["backbone_mask_2d_prev"] = backbone_mask_2d.cpu().numpy()
tmp_dict["backbone_mask_2d_post"] = backbone_mask_2d.cpu().numpy()import pickle
with open("template_pair_embedder_multimer.pkl", "wb") as f:pickle.dump(tmp_dict, f)
logger.info(f"[CL] saved template_pair_embedder_multimer!")
读取特征,即:
def load_tensor_dict(input_path):"""加载特征文件['template_dgram', 'z', 'pseudo_beta_mask', 'backbone_mask', 'multichain_mask_2d','unit_vector_x', 'unit_vector_y', 'unit_vector_z']"""import picklewith open(input_path, "rb") as f:obj = pickle.load(f)print(f"[Info] feat_dict: {obj.keys()}")return objdef process_template_pair_embedder_multimer_dict(feat_dict, output_dir):print(f"[Info] feat_dict.keys: {feat_dict.keys()}")draw_tensor_2d(feat_dict["pseudo_beta_mask_2d_prev"], os.path.join(output_dir, "pseudo_beta_mask_2d_prev.png"))draw_tensor_2d(feat_dict["pseudo_beta_mask_2d_post"], os.path.join(output_dir, "pseudo_beta_mask_2d_prev.png"))draw_template_dgram(feat_dict["template_dgram_post"], os.path.join(output_dir, "template_dgram_post.png"))draw_tensor_2d(feat_dict["backbone_mask_2d_prev"], os.path.join(output_dir, "backbone_mask_2d_prev.png"))draw_tensor_2d(feat_dict["backbone_mask_2d_post"], os.path.join(output_dir, "backbone_mask_2d_post.png"))def draw_tensor_2d(feat, output_path):"""backbone_mask: torch.Size([1, 1102])"""feat = np.squeeze(feat)f, ax_arr = plt.subplots(1, 1, figsize=(8, 5))im = ax_arr.imshow(feat)f.colorbar(im, ax=ax_arr)plt.savefig(output_path, bbox_inches='tight', format='png')plt.show()def draw_template_dgram(feat, output_path):"""template_dgram: torch.Size([1, 1102, 1102, 39])"""f, ax_arr = plt.subplots(6, 7, figsize=(24, 15))ax_arr = ax_arr.flatten()feat = np.squeeze(feat)print(f"[Info] feat: {feat.shape}")for i in range(0, 42):if i <= 38:im = ax_arr[i].imshow(feat[:, :, i], interpolation='none')f.colorbar(im, ax=ax_arr[i])else:ax_arr[i].set_axis_off()plt.savefig(output_path, bbox_inches='tight', format='png')plt.show()
查看全文
99%的人还看了
相似问题
- 【机器学习】特征工程:特征选择、数据降维、PCA
- 论文阅读:“基于特征检测与深度特征描述的点云粗对齐算法”
- 基于纹理特征的kmeas聚类的图像分割方案
- 基于像素特征的kmeas聚类的图像分割方案
- 分类预测 | Matlab实现PSO-GRU-Attention粒子群算法优化门控循环单元融合注意力机制多特征分类预测
- 特征缩放和转换以及自定义Transformers(Machine Learning 研习之九)
- 分类预测 | Matlab实现PSO-BiLSTM-Attention粒子群算法优化双向长短期记忆神经网络融合注意力机制多特征分类预测
- 以key为特征,分类多个信息(手机流量栗子)
- 【机器学习】特征工程:特征预处理,归一化、标准化、处理缺失值
- 004 OpenCV akaze特征点检测匹配
猜你感兴趣
版权申明
本文"PSP - 蛋白质复合物结构预测 Template Pair 特征 Mask 可视化":http://eshow365.cn/6-37682-0.html 内容来自互联网,请自行判断内容的正确性。如有侵权请联系我们,立即删除!